|
Mutation T@sterPhyloP and PhastCons with real data |
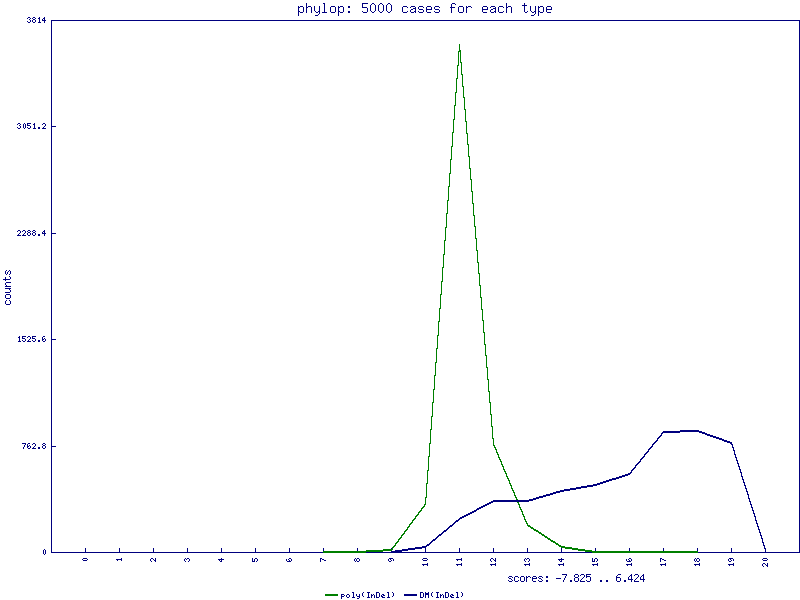
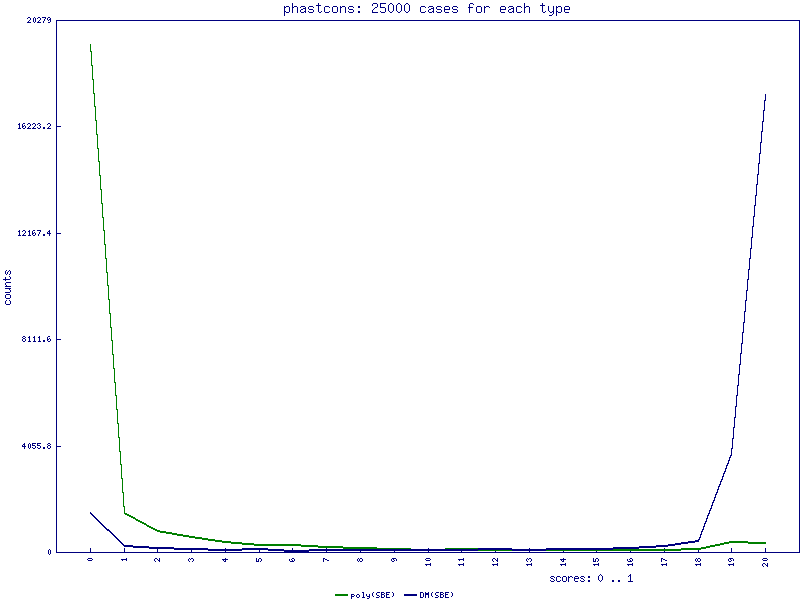
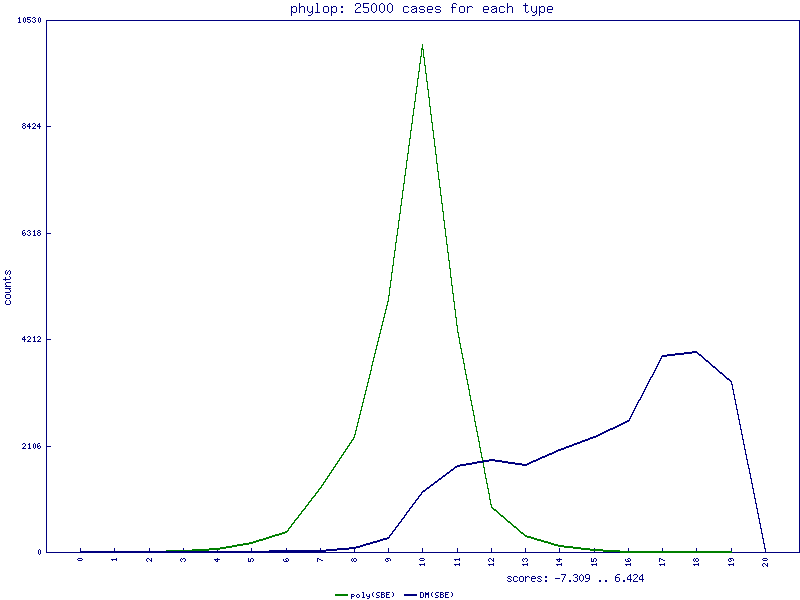
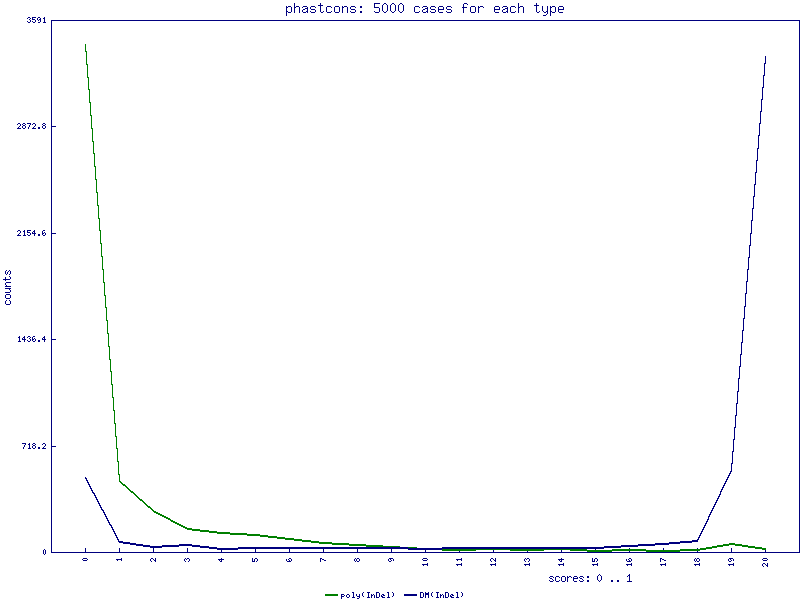
To evaluate the significance of PhyloP and PhastCons scores, we extracted both for a set of
known polymorphisms (from the TGP) and disease mutations (from HGMD Pro).
The outcome is plotted below - with these preliminary results and following further tests,
we decided to include both conservation scores in the classification model as normally distributed variables.
This is, of course, not quite true but gave the best fit in the cross-validation. Using only
the PhastCons scores gave a slightly decreased performance than the combination of both scores. Other tested
alternatives were different classes for each conservation score method, hence the 20 bins plotted
on the X axis.
PhastCons scores for single base exchanges
PhyloP scores for single base exchanges
PhastCons scores for InDels
PhyloP scores for InDels